PathLDM: Text Conditioned Latent Diffusion Model for Histopathology
Abstract
To achieve high-quality results, diffusion models must be trained on large datasets. This can be notably prohibitive for models in specialized domains, such as computational pathology. Conditioning on labeled data is known to help in data-efficient model training. Therefore, histopathology reports, which are rich in valuable clinical information, are an ideal choice as guidance for a histopathology generative model. In this paper, we introduce PathLDM, the first text-conditioned Latent Diffusion Model tailored for generating high-quality histopathology images. Leveraging the rich contextual information provided by pathology text reports, our approach fuses image and textual data to enhance the generation process. By utilizing GPT's capabilities to distill and summarize complex text reports, we establish an effective conditioning mechanism. Through strategic conditioning and necessary architectural enhancements, we achieved a SoTA FID score of 7.64 for text-to-image generation on the TCGA-BRCA dataset, significantly outperforming the closest text-conditioned competitor with FID 30.1.
Model
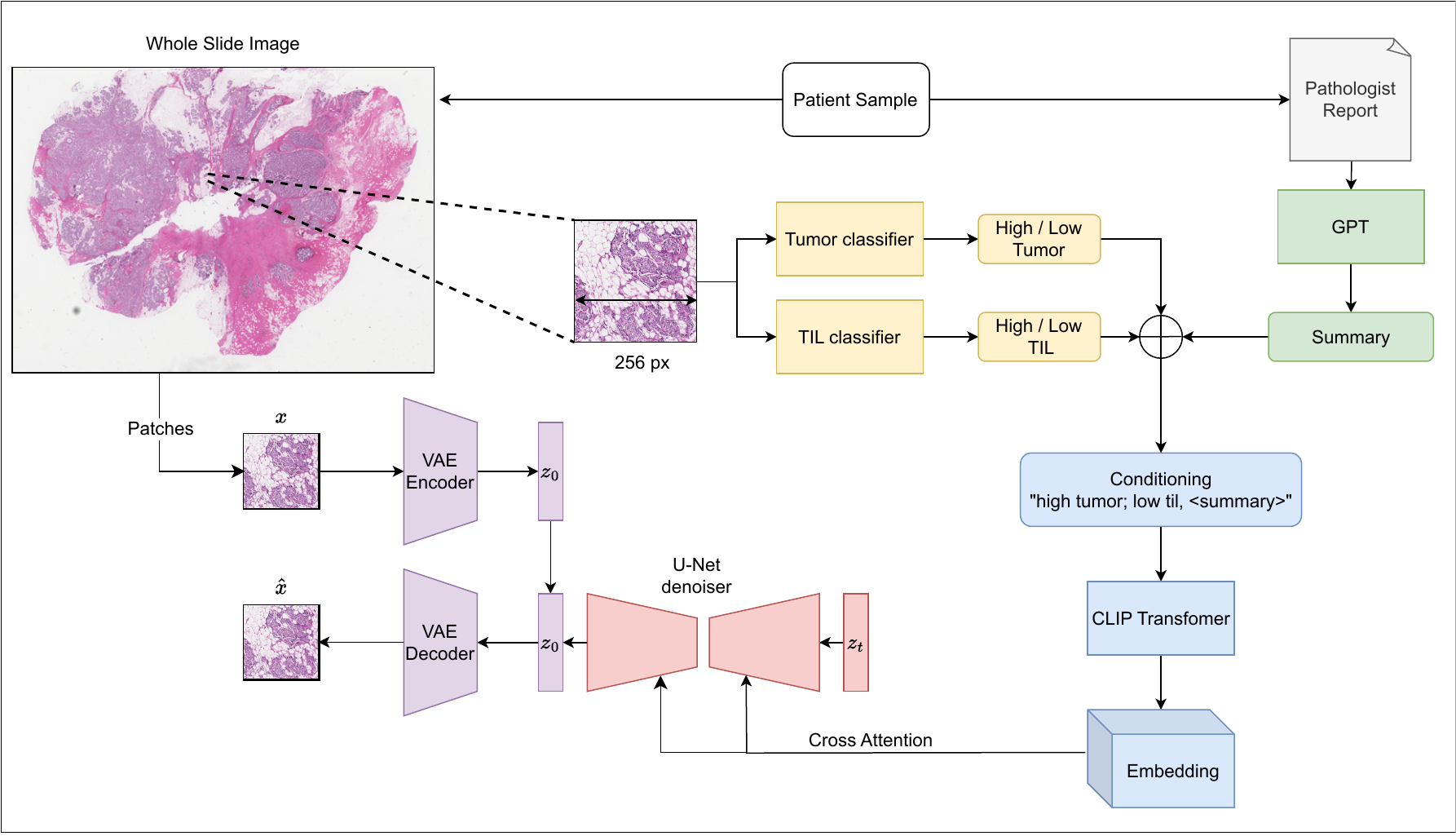
We start with a WSI and an accompanying Pathology report. We crop the WSI into 256 × 256 patches at 10x resolution. Leveraging GPT, we summarize the pathology report. For each patch, we compute Tumor and TIL probabilities, fuse them with the slide-level summary, and condition the LDM with CLIP embeddings of the fused summary. The VAE and text encoder remain frozen, and only the U-Net is trained.
Results
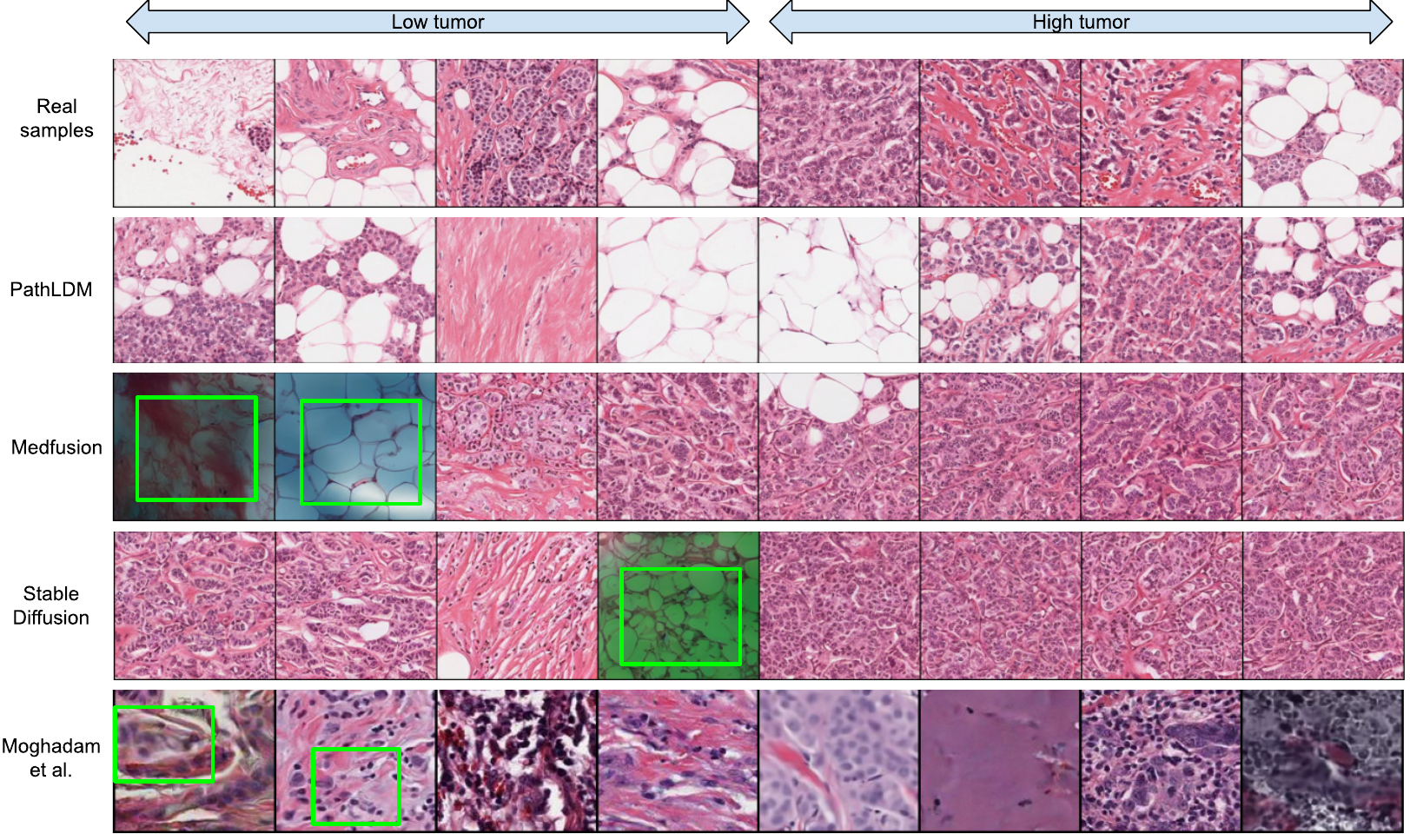
We choose a single text report and produce synthetic samples using Medfusion [29], Stable Diffusion [37], and PathLDM. Samples generated by Medfusion and Stable Diffusion show artifacts (indicated by green boxes) that are not present in our outputs. Moghadam et al. [28] produces images in lower resolution (128 × 128) and exhibits artifacts and blurriness (green boxes in the last row) in the output images. The first row contains the real samples from the corresponding WSI. See paper for citation details.
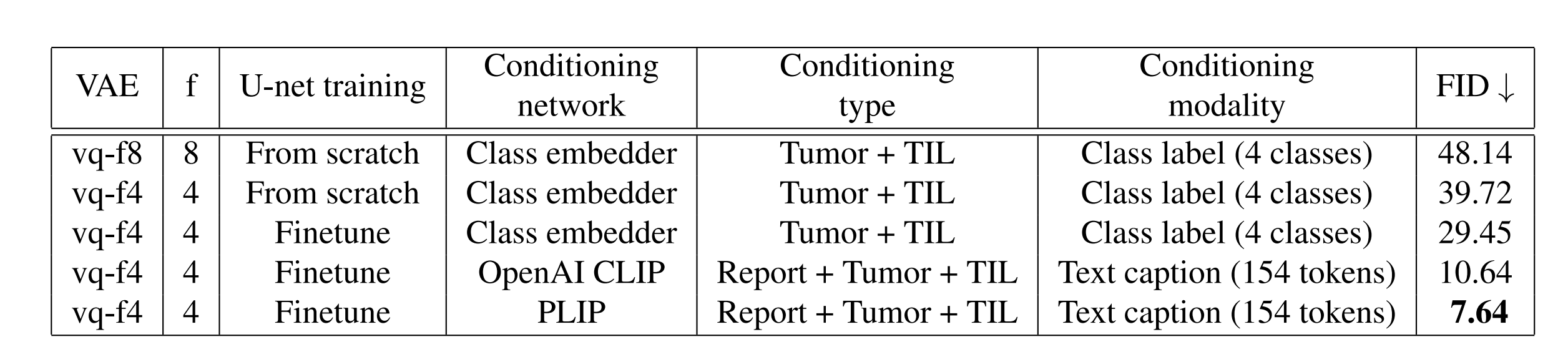
We demonstrate that our effective conditioning, coupled with the architectural refinements, enable PathLDM to achieve a state-of-the-art FID score of 7.64 for text-to-image generation on the TCGA-BRCA dataset.
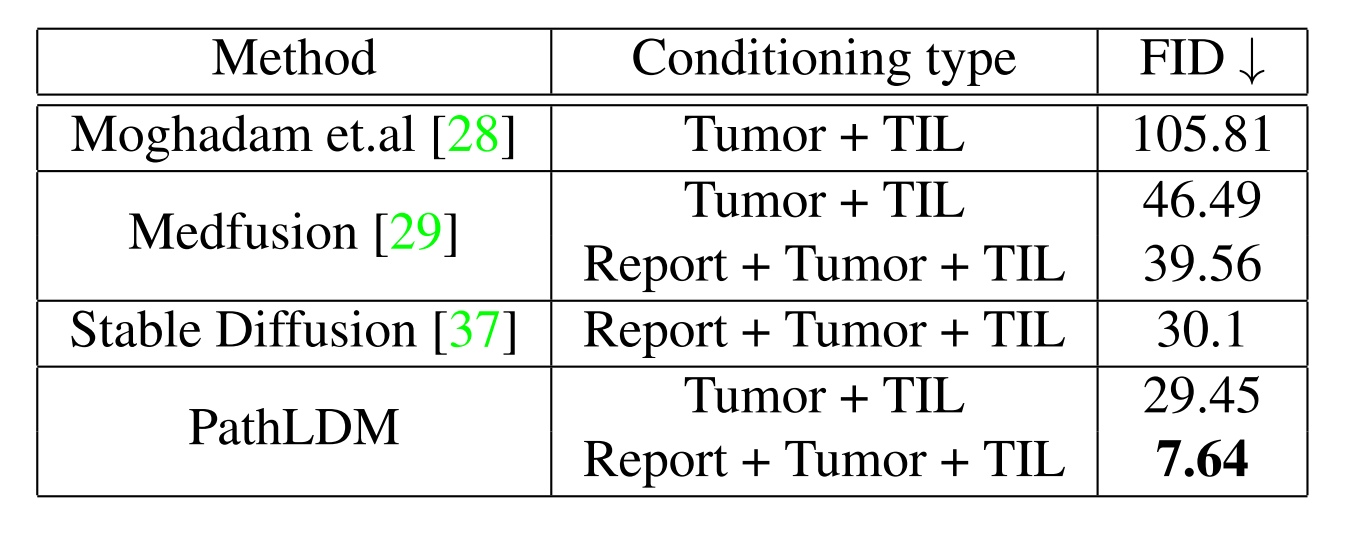
When compared using 10k synthetic images, PathLDM outperforms other methods by a huge margin. Training Medfusion with text-conditioning improved the FID, highlighting the efficacy of our text summaries.
Citation
@InProceedings{Yellapragada_2024_WACV,
author = {Yellapragada, Srikar and Graikos, Alexandros and Prasanna, Prateek and Kurc, Tahsin and Saltz, Joel and Samaras, Dimitris},
title = {PathLDM: Text Conditioned Latent Diffusion Model for Histopathology},
booktitle = {Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV)},
month = {January},
year = {2024},
pages = {5182-5191}
}