PixCell: A generative foundation model for digital histopathology images
Preprint
Srikar Yellapragada*1
Alexandros Graikos*1
Zilinghan Li2
Kostas Triaridis1
Varun Belagali1
Tarak Nath Nandi2,3
Karen Bai1
Beatrice S. Knudsen4
Tahsin Kurc1
Rajarsi R. Gupta1
Prateek Prasanna1
Ravi K. Madduri2,3
Joel Saltz1
Dimitris Samaras1
1Stony Brook University, 2Argonne National Laboratory, 3University of Chicago, 4University of Utah
We present PixCell, the first generative foundation model for digital histopathology. We progressively train our model to generate from 256x256 to 1024x1024 pixel images conditioned on UNI2-h embeddings. PixCell achieves state-of-the-art quality in digital pathology image generation and can be seamlessly used to perform targeted data augmentation and generative downstream tasks.
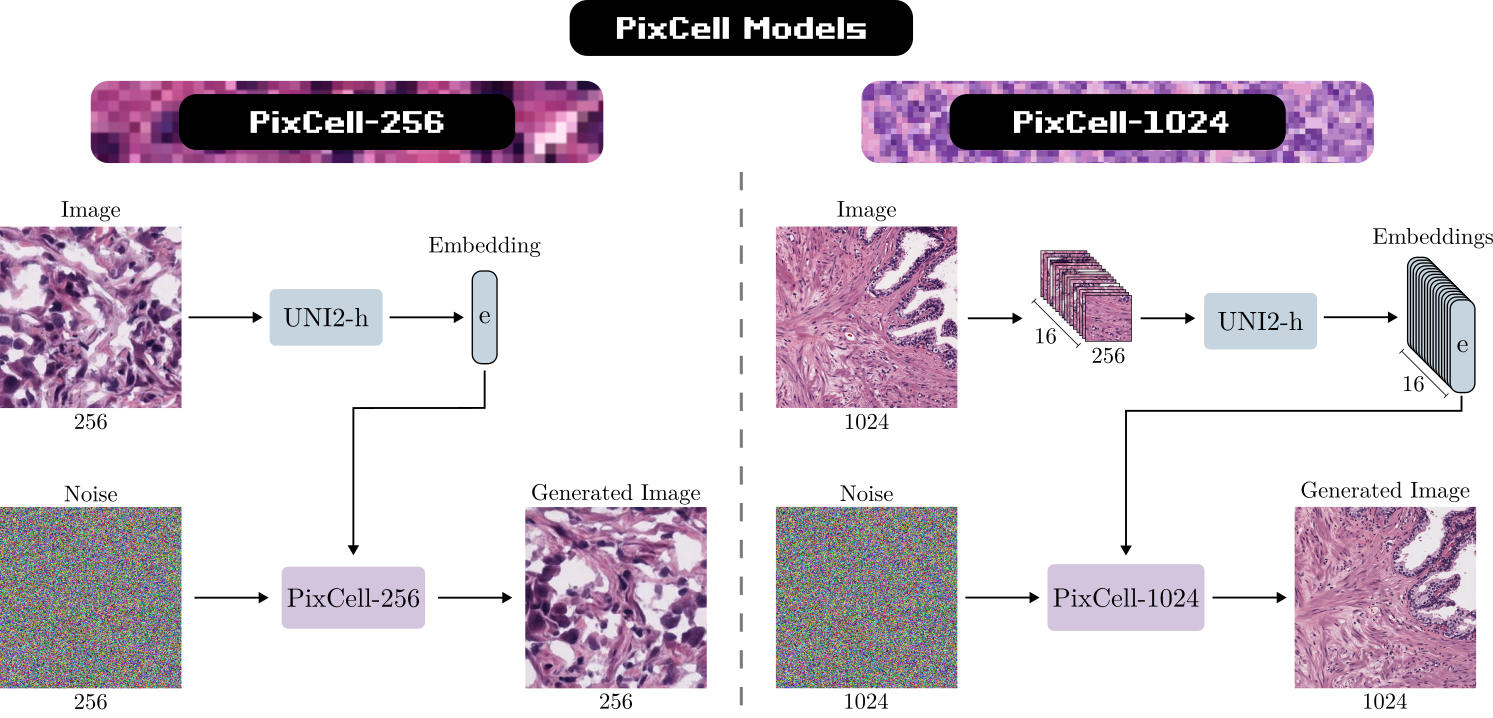
We curate a dataset of 30 million patches at 1024x1024 resolution from public and internal digital histopathology datasets, PanCan-30M. We pair each sample in the dataset with its corresponding 16 UNI-2h embeddings to construct the training data. We progressively train a diffusion transformer model, starting from 256x256 resolution and scaling up to 1024x1024. We release the weights for both the 256x256 model (PixCell-256) and the 1024x1024 model (PixCell-1024).
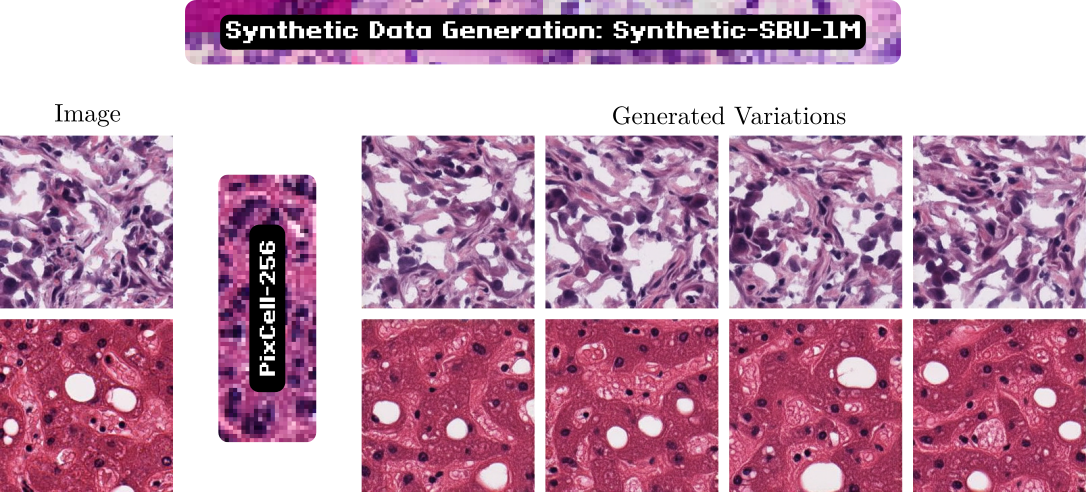
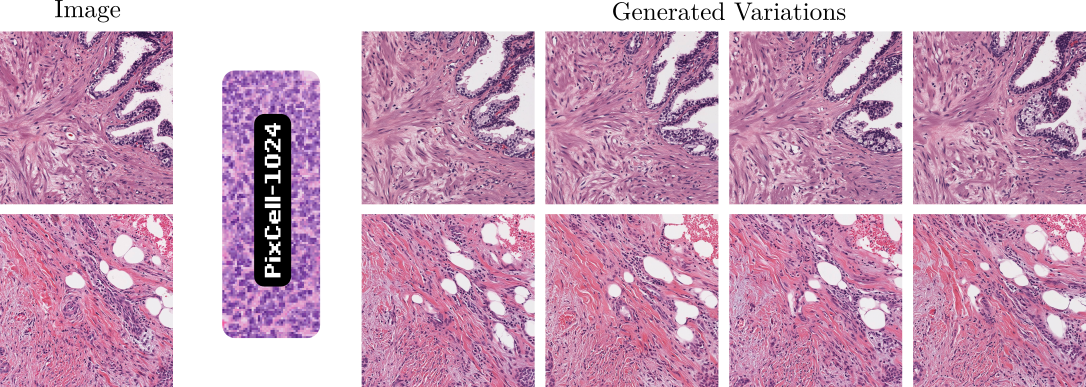
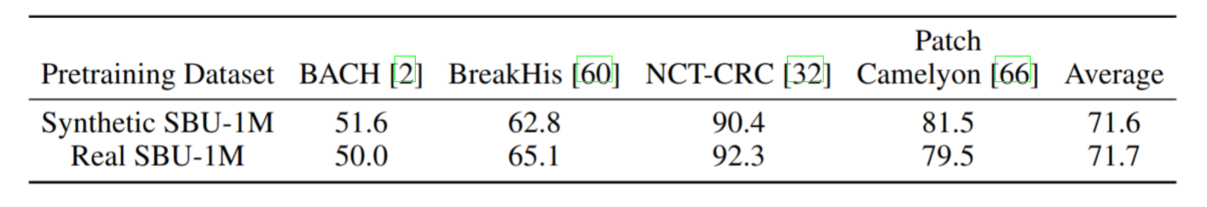
Given a UNI embedding from a reference image, PixCell generates images with similar appearance and content. Using the PixCell-256 model we generate a synthetic variant of our SBU-1M dataset, Synthetic SBU-1M. We show that we can train an encoder on the synthetic data only without losing any performance on downstream tasks.
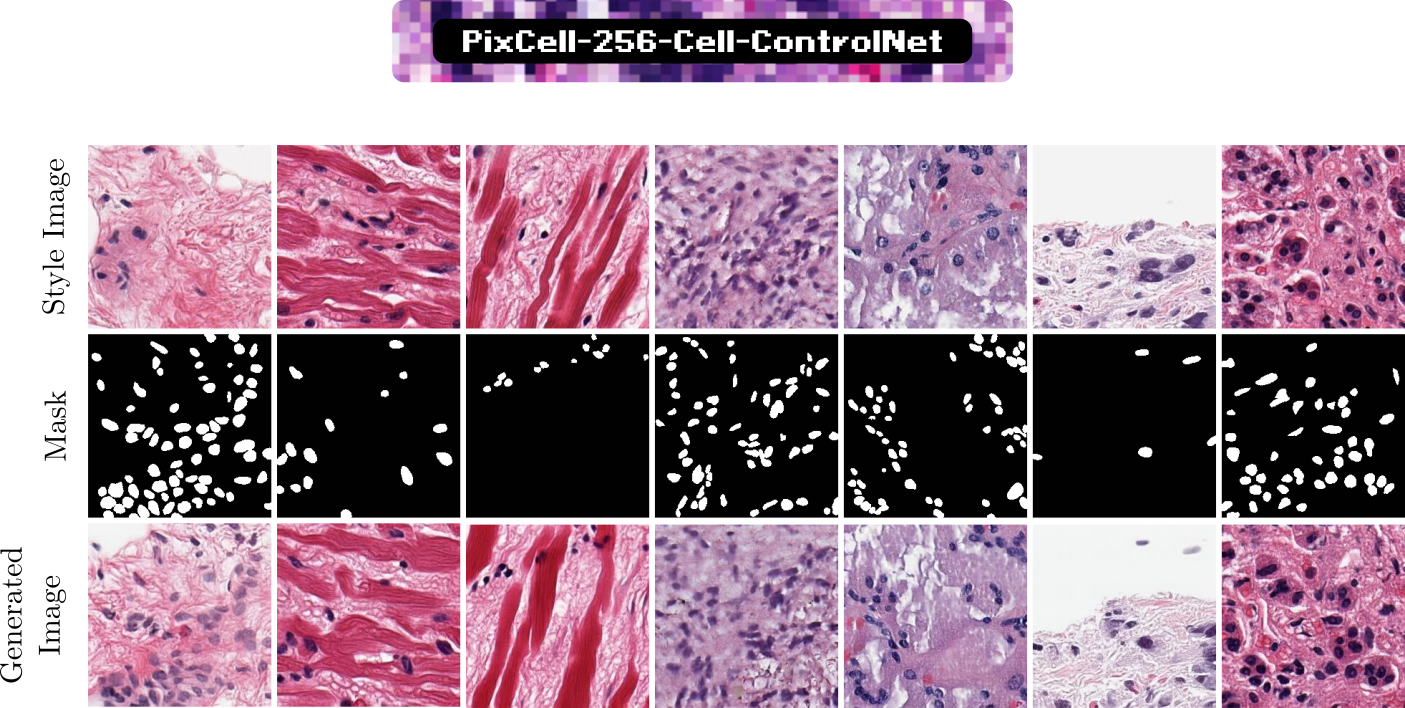
Using a pre-trained cell segmentation model, we construct a dataset of 10,000 image-cell mask pairs. We train a cell mask ControlNet (PixCell-256-Cell-ControlNet) on this dataset to guide image generation. Using the UNI embedding from a reference image to control the style and a cell mask to control the layout, we can generate targeted synthetic data using the appearances from the test set and masks from training set to improve the downstream cell segmentation task.
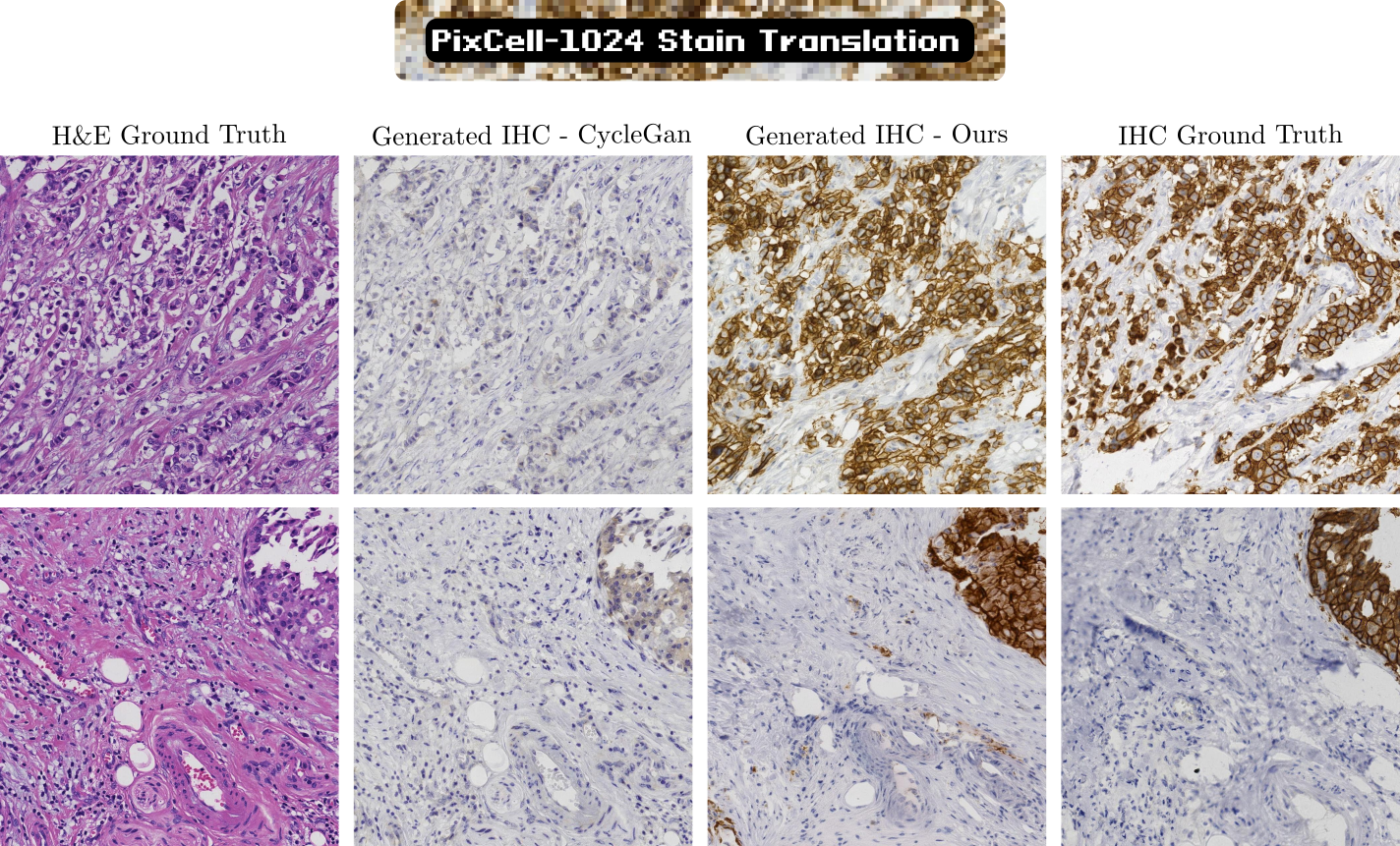
Our PixCell models, although never trained explicitly on IHC-stained data, can generalize to IHC images. Using a dataset of "roughly" paired H&E and IHC patches we learn a transformation between H&E UNI embeddings and IHC UNI embeddings. We use this learned transformation to perform stain translation between the two different staining techniques, significantly outperforming previous GAN-based models.
Citation
@article{yellapragada2025pixcell,
title={PixCell: A generative foundation model for digital histopathology images},
author={Yellapragada, Srikar and Graikos, Alexandros and Li, Zilinghan and Triaridis, Kostas and Belagali, Varun and Kapse, Saarthak and Nandi, Tarak Nath and Madduri, Ravi K and Prasanna, Prateek and Kurc, Tahsin and others},
journal={arXiv preprint arXiv:2506.05127},
year={2025}
}