Pathology Image Compression with Pre-trained Autoencoders
TL;DR
Large volumes of high-resolution digital histopathology whole-slide images are essential in developing large-scale machine learning models. However, storage, transmission, and computational efficiency are significant challenges that need to be overcome in order to efficiently utilize these vast repositories. We find that existing autoencoders are better pathology image compressors than JPEG and propose to use them to reduce the costs of storing large whole-slide image repositories.
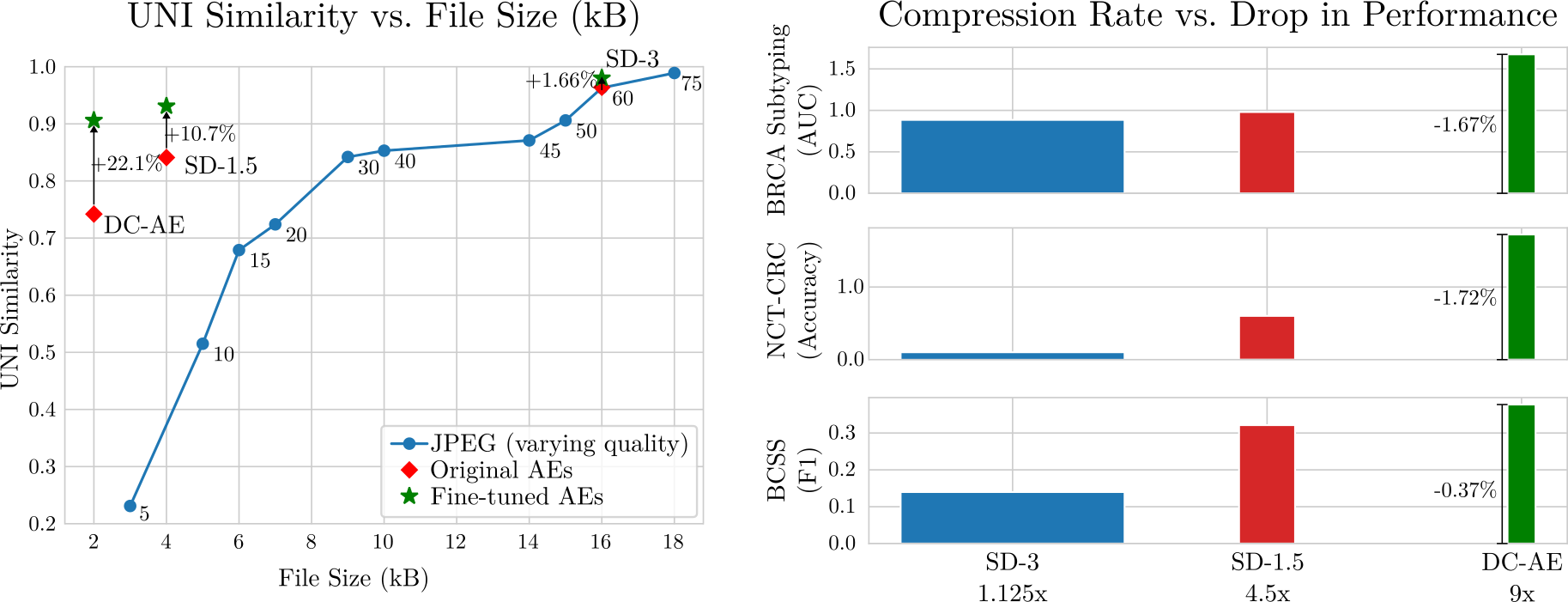
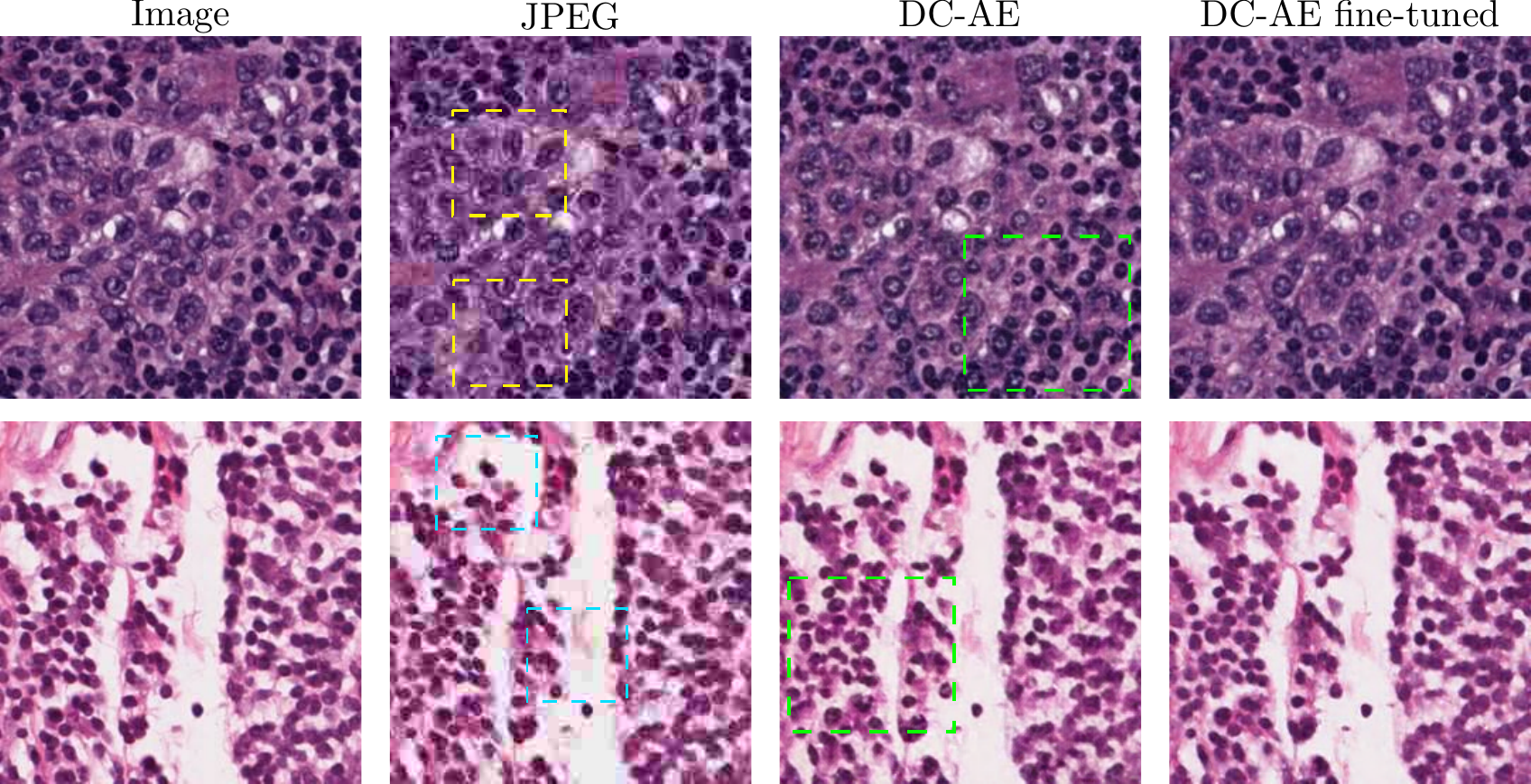
Furthermore, in cases where the AEs fail to preserve fine-grained phenotypic details, we show that fine-tuning only the decoder of existing AE models with a pathology-specific perceptual metric increases the quality of the reconstructions significantly.
Using compressed images in downstream tasks
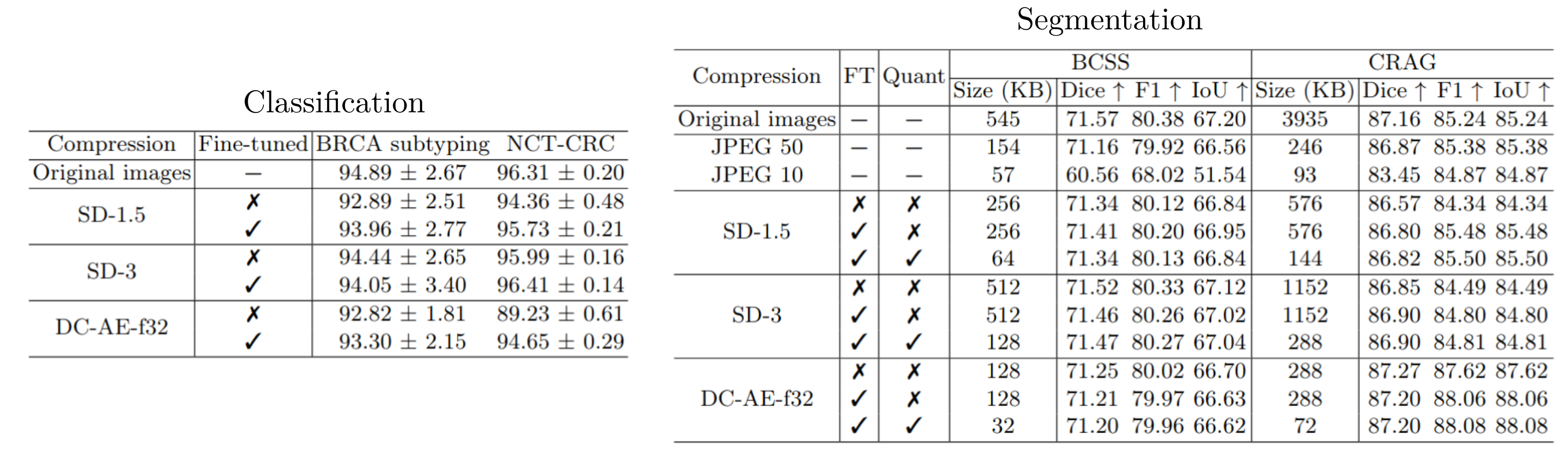
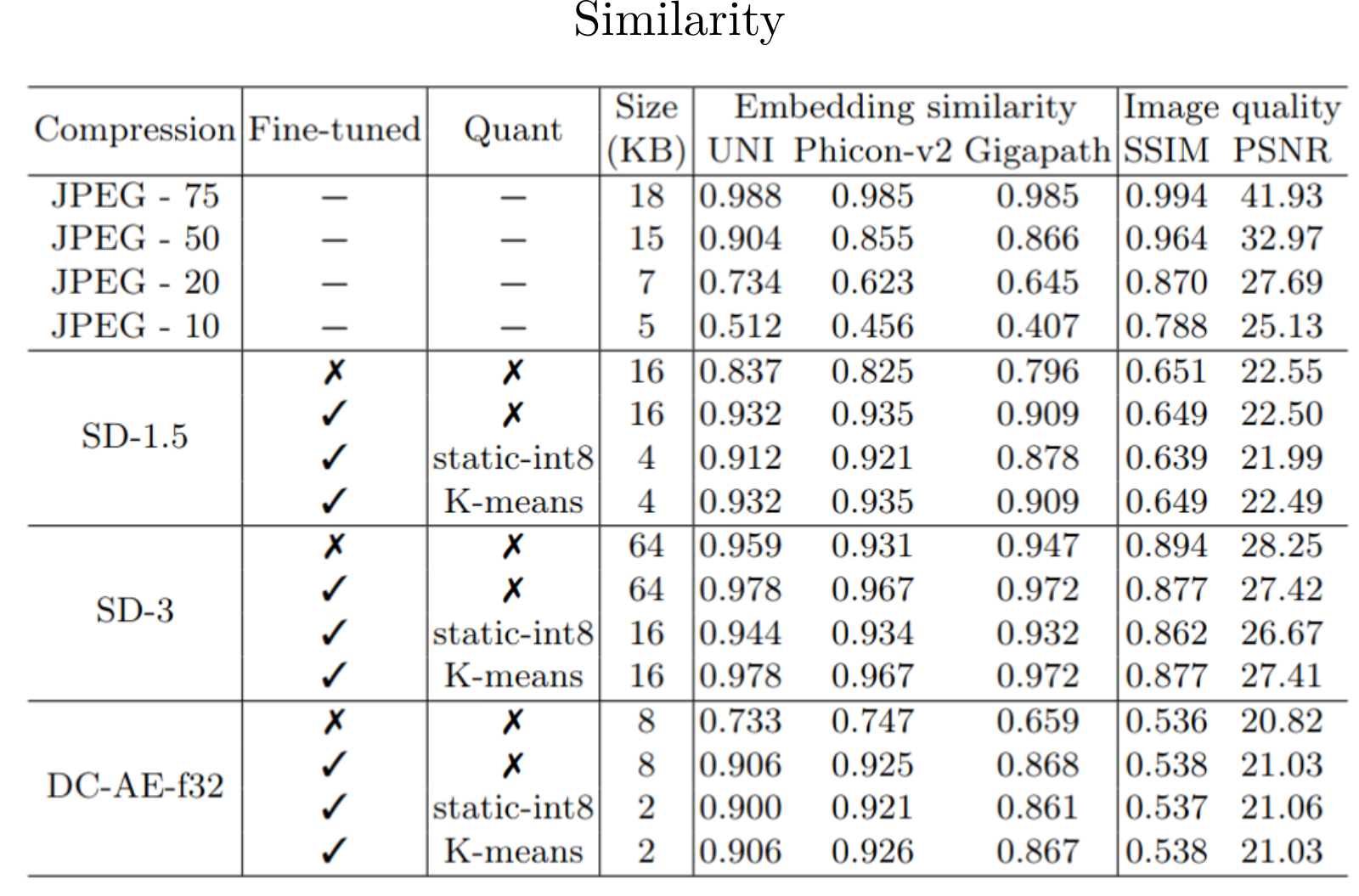
We systematically benchmark three autoencoders with varying compression levels on segmentation, patch classificationand and multiple instance learning by replacing the original images with their autoencoder reconstructions. Using AE-compressed images leads to minimal performance degradation. Employing a K-means clustering-based quantization method for the autoencoder latents, we reduce storage requirements by as much as 8x.
Citation
@article{yellapragada2025pathology,
title={Pathology Image Compression with Pre-trained Autoencoders},
author={Srikar Yellapragada and Alexandros Graikos and Kostas Triaridis and Zilinghan Li and Tarak Nath Nandi and Ravi K Madduri and Prateek Prasanna and Joel Saltz and Dimitris Samaras},
journal={arXiv preprint arXiv:2503.11591},
year={2025},
}